Create S2 L1C Sample Data¶
A S2 L1C (S2A_MSIL1C_20170216T102101_N0204_R065_T33UUU_20170216T102204.SAFE) scene downloaded from the Copernicus Open Access Hub comes as several individual JPEG2000 raster files which have different resolution. In this notebook we clip and save a small window from these layers by keeping the resolution intact. The data created here is contained as sample and test dataset s2l1c in the eo-box package.
Let’s first import the packages we need:
[2]:
from affine import Affine
import glob
import matplotlib.pyplot as plt
import os
import pandas as pd
import rasterio
from rasterio.windows import Window
import seaborn as sns
% matplotlib inline
Now we need the layer files and some information from the full-tile S2 L1C scene.
[5]:
scene_dir = r'xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0204_R065_T33UUU_20170216T102204.SAFE'
layer_files = glob.glob(f'{scene_dir}/**/*_[B]??.jp2', recursive=True)
parts = [os.path.basename(file).split(".")[0].split("_") for file in layer_files]
layers_df = pd.DataFrame(parts, columns=["tile", "datetime", "band"])
layers_df["file"] = layer_files
layers_df = layers_df.set_index("band").sort_index()
layers_df
[5]:
| tile | datetime | file | |
|---|---|---|---|
| band | |||
| B01 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B02 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B03 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B04 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B05 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B06 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B07 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B08 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B09 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B10 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B11 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B12 | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
| B8A | T33UUU | 20170216T102101 | xxx_uncontrolled/S2A_MSIL1C_20170216T102101_N0... |
We want to clip the same small spatial area from all layers. Let’s get the window from one of the 60m layers.
[6]:
win_60m = Window(col_off=500, row_off=1300, width=2**8, height=2**7)
with rasterio.open(layers_df.at["B01", "file"]) as src:
meta_60m = src.meta
arr_60m = src.read(1, window=win_60m)
fig, ax = plt.subplots(1, 1, figsize=(16, 8))
sns.heatmap(arr_60m, robust=True, cbar=False)
[6]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f4985dc9748>
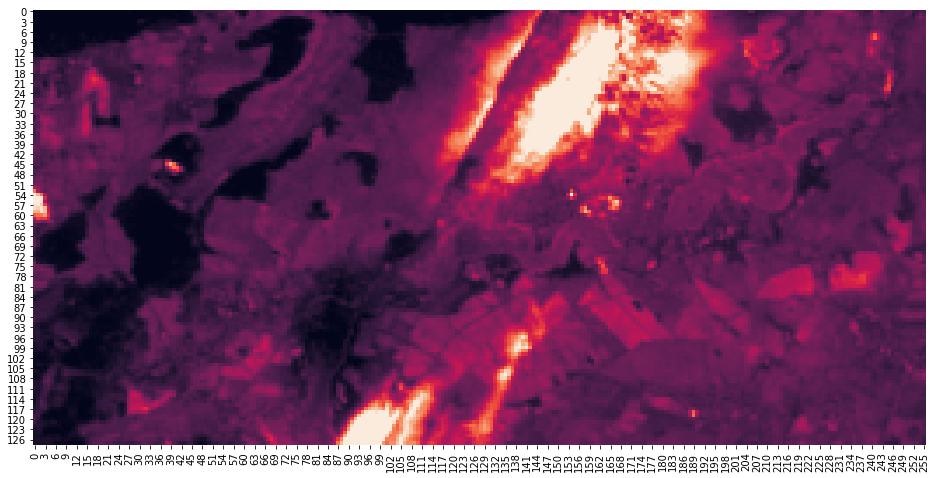
Since the layers have all different resolution / cell size, we need to derive the corresponding windows for the other layer. This is implemented in the follwoing function.
[7]:
def transform_window(window_src, transform_src, transform_dst):
spatial_bounds = rasterio.windows.bounds(window=window_src, transform=transform_src,
height=0, width=0) # defaults
window_dst = rasterio.windows.from_bounds(spatial_bounds[0],
spatial_bounds[1],
spatial_bounds[2],
spatial_bounds[3],
transform=transform_dst,
height=None, width=None, precision=None) # defaults
return window_dst
Visually and with respect to the window relations the function seems to do what it should.
[8]:
with rasterio.open(layers_df.at["B02", "file"]) as src:
meta_10m = src.meta
win_10m = transform_window(window_src=win_60m,
transform_src=meta_60m["transform"],
transform_dst=meta_10m["transform"])
arr_10m = src.read(1, window=win_10m)
assert(win_10m.height == win_60m.height * 6)
assert(win_10m.width == win_60m.width * 6)
assert(win_10m.row_off == win_60m.row_off * 6)
assert(win_10m.col_off == win_60m.col_off* 6)
fig, ax = plt.subplots(1, 1, figsize=(16, 8))
sns.heatmap(arr_10m, robust=True, cbar=False)
[8]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f498370b0f0>
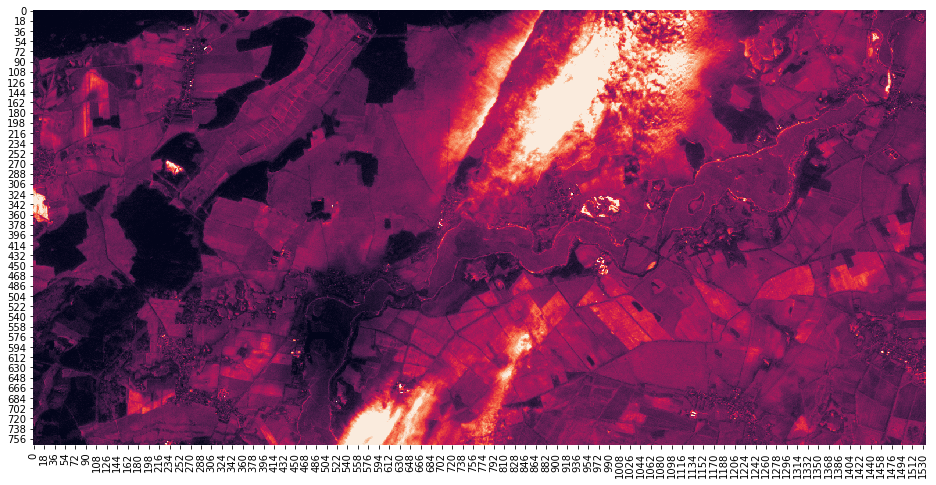
Now we can loop through all the layers and save the subset. The destination files of the subsetted files are printed out.
[13]:
dst_dir = r'xxx_uncontrolled/s2l1c'
# from the 60 m
win = Window(col_off=500, row_off=1300, width=2**8, height=2**7)
transform = Affine(60.0, 0.0, 300000.0, 0.0, -60.0, 5900040.0)
blockxsize = blockysize = 2**6
for filename_src in layers_df["file"].values:
filename_dst = os.path.join(dst_dir, *filename_src.split("/")[-2:])
bprint(filename_dst)
if not os.path.exists(os.path.dirname(filename_dst)):
os.makedirs(os.path.dirname(filename_dst))
if not os.path.exists(filename_dst):
with rasterio.open(filename_src) as src:
win_this = transform_window(window_src=win,
transform_src=transform,
transform_dst=src.meta["transform"])
arr = src.read(window=win_this)
kwargs = src.meta.copy()
kwargs.update({
'height': win_this.height,
'width': win_this.width,
'transform': rasterio.windows.transform(win_this, src.transform),
'compress': "JPEG2000"})
with rasterio.open(filename_dst, 'w', **kwargs,
tiled=True, blockxsize=blockxsize, blockysize=blockysize) as dst:
dst.write(arr)
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B01.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B02.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B03.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B04.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B05.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B06.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B07.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B08.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B09.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B10.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B11.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B12.jp2
xxx_uncontrolled/s2l1c/IMG_DATA/T33UUU_20170216T102101_B8A.jp2
The End.